
Yvonne TAY
In recent years, next-generation sequencing has revealed the existence of thousands of non-coding RNAs, which comprise the majority of the human transcriptome. Our work focuses on understanding the role that these non-coding RNAs play in cancer development, and exploring their potential utility as diagnostic biomarkers and targeted therapeutics for precision medicine.
csitmsy[at]nus.edu.sg
Principal Investigator, Cancer Science Institute of Singapore, NUS
Associate Professor, Department of Biochemistry, NUS.
| 2022, 2021 | Faculty of Dentistry Excellence in Teaching Award |
| 2021 | Yong Loo Lin School of Medicine Teaching Excellence Award |
| 2021 | Yong Loo Lin School of Medicine Young Researcher of the Year Award |
| 2015 | Young Scientist Award |
| 2015 | Singapore National Research Foundation Fellowship |
| 2014 | NUS President’s Assistant Professorship |
| 2011-2014 | Special Fellowship, Leukemia & Lymphoma Society |
| 2009 | Philip Yeo Prize for Outstanding Achievement in Research, A*STAR Biomedical Research Council, Singapore |
| 2004-2008 | A*STAR Graduate Scholarship, Agency for Science, Technology and Research, Singapore |
The dysregulated expression of critical genes is one of the driving forces that transforms a normal cell into a cancer cell. In addition to genomic alterations, aberrant changes in post-transcriptional regulation by factors including non-coding RNAs and RNA binding proteins represent another mechanism to modify gene function and thus contribute to tumorigenesis.
The advent of next-generation sequencing technologies has led to the identification of thousands of non-coding RNAs, only a handful of which have been functionally characterized. In addition to studying non-coding RNAs in their own right, our group is also interested in studying the non-coding regions of protein-coding mRNAs (untranslated regions, UTRs). As many mRNA populations comprise transcripts with different UTRs, and these UTRs control key processes such as stability, localization and transport, a better understanding of their function may lead to insights into the regulation of key cancer genes.
Our work has three main focus areas: (1) Deconvoluting RNA:RNA networks in cancer and understanding how they contribute to carcinogenesis; (2) Understanding the interplay between RNA:RNA interactions and RNA:protein interactions and (3) Examining potential crosstalk between these post-transcriptional networks and RNA processing pathways such as splicing and editing. Our long-term goal is to translate our basic research discoveries into the clinic, we anticipate that our work will open new avenues for the development of novel RNA-based anti-cancer diagnostics and therapeutics.
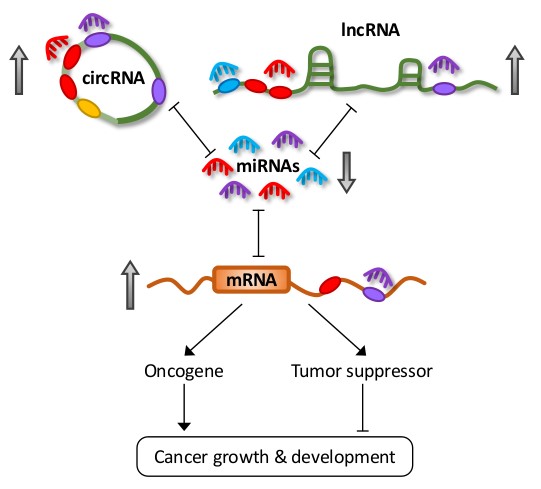
Fig 1. Different species of RNAs, including messenger RNAs (mRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) can co-regulate each other in competing endogenous RNA networks by sequestering or ‘sponging’ shared microRNAs. The dysregulation of any single RNA can lead to a ripple effect across the network and thus modulate carcinogenesis.
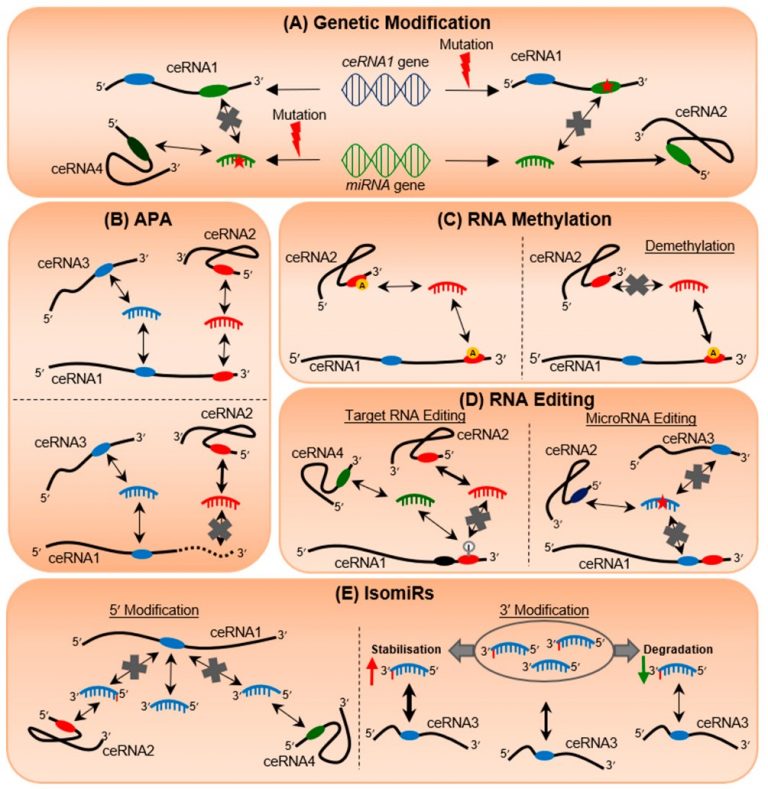
Fig 2. The butterfly effect of RNA alterations on transcriptomic equilibrium. The different modifications affecting either target RNAs or miRNAs and/or both, such as genetic modification (A), alternative polyadenylation (APA) (B), RNA methylation (C), RNA editing (D), and isomiR production (E). Ovals represent the miRNA response elements (MREs); double-headed arrows represent bi-directional regulation in which the thickness indicates the strength of the regulation; single-headed arrows indicate increase (pointed up) or decrease (pointed down) in RNA expression; red stars indicate non-specific nucleotide alteration; grey crosses indicate loss of interaction; red lightning bolts represent mutation events; yellow circles with an A represent methylation marks; grey circle with an I represents A-to-I RNA editing.








