Quantitative Proteomics Core
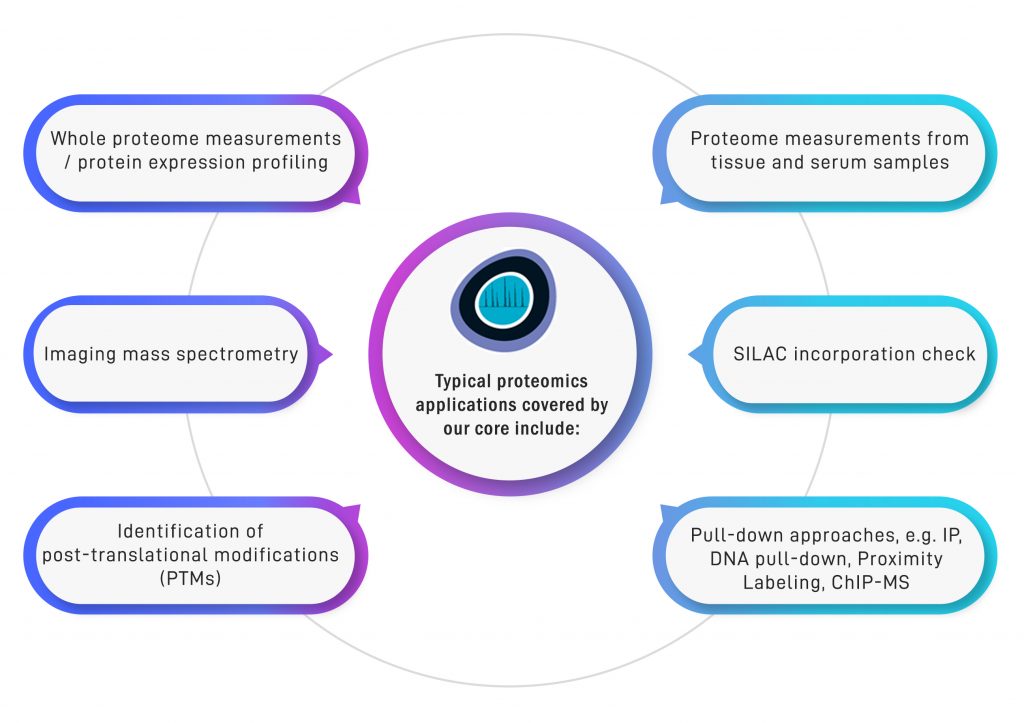
The Quantitative Proteomics Core at the Cancer Science Institute of Singapore offers a complete package for proteomics applications all the way from help with experimental design over sample preparation and mass spectrometry measurement to data analysis, visualization and storage. We cover a broad range of proteomics applications including clinical samples.
We currently operate a Q Exactive HF hybrid quadrupole-Orbitrap mass spectrometer as well as a timsTOF fleX in combination with an EASY-nLC 1200 uHPLC system or an Evosep ONE. The data analysis is performed on an in-house server using the freely available quantitative proteomics software package MaxQuant. We are committed to providing quantitative mass spectrometry solutions and to this end support researchers in using SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture) or label-free quantitation (LFQ).
For further information and to discuss how we may help you with your specific experimental questions, please contact the Quantitative Proteomics Core via mail (csi_proteomics[at]nus.edu.sg).
Facility Head

Dr. Dennis KAPPEI
CSI Singapore
dennis.kappei[at]nus.edu.sg
Team

Vartika KHANCHANDANI
Research Assistant
Vartika earned a Bachelor of Engineering from the R.V. College of Engineering in Bangalore. In our group, she is applying her bioinformatics skills to the development of streamlined analysis pipelines.

