Yoshiaki ITO
Professor Ito’s research centers on the regulation of stem cells in the stomach. Stem cells are required for daily maintenance of the stomach, which is constantly damaged by microorganisms and chemicals. The discovery of RUNX genes in 1993 led the Ito group to uncover the dual role of RUNX3 – both as a tumour suppressor and an oncogene. More recently in 2017, RUNX1, a known stem cell gene in hematopoiesis, was also found to be essential for proliferating stem cells in the stomach. This finding led us to identify stem cell specific protein, IQGAP3, as functionally important for homeostasis, tissue repair and cancer in 2020. IQGAP3 is an embryonic protein which is expressed in rapidly proliferating cancer cells in virtually all types of cancer and knockdown of IQGAP3 induces differentiation of cancer cells. Cancer is like a wound that does not heal. Professor Ito’s group is currently studying the requirement of IQGAP3 in the process of wound healing and cancer development.
csiitoy[at]nus.edu.sg
Senior Principal Investigator, Cancer Science Institute of Singapore, National University of Singapore
Yong Loo Lin Professor of Medical Oncology, Department of Medicine, Yong Loo Lin School of Medicine, NUS.
| 2010 | President Science Award, Singapore |
| 2003 | Tomizo Yoshida Prize, Japanese Cancer Association |
| 1995 | Princess Takamatsu Cancer Research Award, Princess Takamatsu Cancer Research Foundation, Japan |
| 1968 | Kuroya Award, Japanese Society for Microbiology |
Historical flow of our research activities from RUNX to stem cells:
In 1993, three groups including my own reported a new class of transcription factor family, now called the RUNX family [Trends Genet 9:338-41,1993]. RUNX genes encode transcription factors that function as master developmental regulators. They are also frequently involved in cancer. Since their discovery, our group has been studying the roles of the RUNX family, especially RUNX3, in carcinogenesis. We found that RUNX genes have paradoxical roles, functioning as tumor suppressors or oncogenic drivers, depending on the cell context [Nat Rev Cancer 15:81-95, 2015].
Aside from transcription regulation, we have also uncovered unexpected links of RUNX3 protein to the Fanconi anemia DNA repair pathway, TGFb-induced genomic instability and Aurora kinases during mitotic entry – all of which support the role of RUNX3 as a primary line of defense against oncogenic stimuli [Cell Rep 24:1747-55, 2018; PNAS 113:6490-5, 2016; Cancer Res 78:88-102, 2018; Cell Rep 8:767-82, 2014].
Using mouse models, we found that knockout of RUNX3 induces adenoma in the intestine, lung, mammary gland as well as precancerous lesions in the stomach [Oncogene 29:2605-15, 2010; Gastroenterol 140:1536-46, 2011; Cancer Cell 14:226-237, 2008]. This is the cornerstone of our argument that RUNX3 is a tumor suppressor and gatekeeper of cancer development. RUNX3 can also function as an oncogene. We have an excellent tool to solve this puzzle. We found that a single nucleotide change R122C in RUNX3, identified in a patient of gastric cancer, results in its conversion from tumor suppressor to oncogene [Cell 109: 113-24, 2002]. By introducing this single nucleotide change in the mouse genome, we are finding that RUNX3 has an important role in the cell cycle regulation in the adult stem cells.
Recently, an exciting new concept of RUNX as a key regulator of adult stem cells in multiple organs has emerged. A 270 bp element of RUNX1 enhancer, termed eR1 [Stem Cells 28: 1869-81, 2010], is responsible for the specific expression of RUNX1 in hematopoietic stem cells. We subsequently found that eR1 also drives the expression of RUNX1 in tissue stem cells of multiple organs. Having developed eR1 as a powerful tool to study stem cells, we identified the elusive rapidly proliferating stomach isthmal epithelial stem cells [Gastroenterol 152:218-231, 2017]. We explored the protein and gene expression profiles of these isthmal stem cells and found strong enrichment of a cytoskeletal scaffold protein IQGAP3 (IQ motif containing GTPase activating protein 3) [GUT 2020 doi:10.1136/gut jnl-2020-322779]. This scaffold protein interacts with multiple proteins to enhance signaling intensity. Among them, the GTP bound form of Ras is particularly important. Furthermore, we found that IQGAP3 is expressed in the majority, if not all, of rapidly proliferating gastric cancer cells. A comparison with the expression of cancer stem cell marker indicated that IQGAP3 expression may mark a rapidly proliferating population of cancer stem cells. The expression of IQGAP3 in multiple cancer types indicates a broad role for IQGAP3 in oncogenesis.
We are also studying how IQGAP3 is specifically expressed in stem cells and how IQGAP3 maintains stem cell property. Promising regulators of IQGAP3, including an RNA binding protein, IGF2BP1, have been identified. Depletion of IQGAP3 or its upstream regulators in stem cells or gastric cancer cells induced cellular differentiation, accompanied by sharp decreases of transcription of stem cell factors such as Oct4, Nanog and Sox2. IQGAP3 therefore has a capability to maintain the transcriptional signature of stem cells. This induction of differentiation in aggressive, poorly differentiated cancer cells indicates a novel area for therapeutic exploitation – development of inhibitors against IQGAP3 or its upstream regulators may lead to effective reprogramming of proliferating cancer stem cells and activation of the terminal differentiation process.
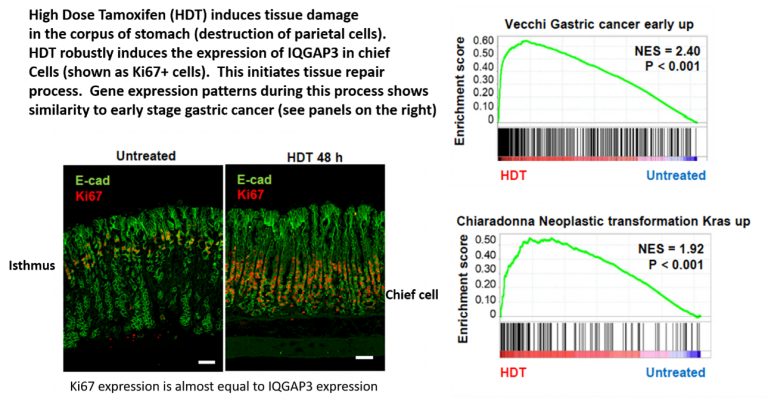
Another important aspect of IQGAP3 is its role in tissue repair. Soon after tissue damage, IQGAP3 is robustly induced in normally quiescent, fully differentiated zymogenic chief cells. This induction is associated with the reprogramming of chief cells, which acquire proliferative and stem-like properties. Furthermore, transcriptomic analysis of the damaged tissues revealed striking similarities to the gene expression signature of early gastric cancer. There is a classical observation that cancer is ‘a wound that does not heal’ [N Engl J Med 315:1650-9, 1986]. Therefore, coupled with the fact that IQGAP3 is strongly expressed in cancer cells, we propose that the IQGAP3-driven repair process is linked to the carcinogenic process and that we are observing the beginnings of cancer.