Wee Joo CHNG
Professor Chng Wee Joo’s research focus is on the use of global genomics methods (microarray and sequencing platforms) to study the clinical and biological heterogeneity in haematologic malignancies including acute myeloid leukemia, multiple myeloma and lymphoma. Using these methods, he has identified novel prognostic markers as well as molecular abnormalities in these malignancies, thereby providing insights into disease pathogenesis and biology, and serve as potential targets for therapy.
mdccwj[at]nus.edu.sg
Senior Principal Investigator, Cancer Science Institute of Singapore, NUS
Yong Loo Lin Professor in Medical Oncology, Yong Loo Lin School of Medicine, NUS
Vice President (Biomedical Sciences Research), Office of the Deputy President (Research and Technology), NUS
Group Chief Scientist, National University Health System
Senior Consultant, National University Cancer Institute, Singapore, National University Health System
Executive Director, Singapore Translational Cancer Consortium (STCC)
To list just a few of the many accolades received:
|
2024 |
Distinguished Senior Clinician Award (DSCA) |
|
2024 |
NMRC 2024 Singapore Translational Research Investigator Award |
|
2023 |
COVID19 Resilient Medal |
|
2022 |
NUHS Pinnacle Award - Research |
|
2021 |
School of Medicine Graduate Mentor of the Year Award |
|
2020 |
Brian G.M. Durie Outstanding Achievement Award |
|
2020 |
Public Administration Medal (Silver) |
|
2018 |
NUS YLL SOM Provost’s Chair Professorship |
|
2017 |
NUS YLL SOM Outstanding Researcher of the Year Award |
|
2017 |
NMRC Senior Translational (STaR) Research Award |
|
2016 |
26th Seah Cheng Siang Memorial Lecture |
|
2016 |
NMEA National Outstanding Clinician-Scientist Award |
|
2016 |
NUHS Academic Medicine Development Award |
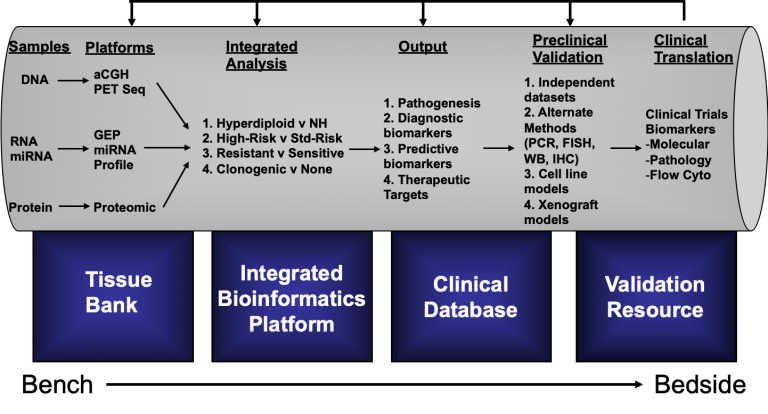
The Haematological Malignancy Genomics Laboratory operates a comprehensive translational research program in haematological malignancies with focus on multiple myeloma (MM), acute myeloid leukemias (AML) and natural killer / T-cell Lymphoma (NKTL). At the core of this program is the use of high-throughput cutting edge genomics and proteomics techniques in human tumor samples and model system to make clinically relevant discoveries. These discoveries will encompass novel biological insights, identification of new diagnostic subtypes, prognostic factors, therapeutic targets, and aspects of molecular epidemiology and pharmacogenomics, all with potential impact on patient care. In this bench-to-bedside translational pipeline, discoveries are validated in the pre-clinical setting before clinical validation. The program will be supported by a comprehensive tissue bank that provides high-quality source materials for down-stream study, and a clinical database that is connected by a relational database for integrated system biology analysis (See Figure Below). In MM, we are focused on identifying the pathways leading to disease progression. In this regard, we have constructed step-wise pathways of progression for MM and is using this as a framework to design therapeutic intervention strategy. At the same time, we are using genomics to dissect the molecular heterogeneity of the disease. This has yielded robust genetic subtypes. We are now focusing on rationally targeting high-risk subtypes based on the underlying genomic aberrations and molecular defects. In AML, we have been working on mechanisms mediating therapeutic resistance in FLT3 positive AML, and in the process have identified novel molecules that play a fundamental role in leukemogenesis and may represent novel therapeutic targets. In addition, we are also testing novel compounds targeting EZH2, an oncogenic histone modifier, and has unravelled interesting biology in AML so far. In NKTL, we are using genomics to understand key molecular event mediating pathophysiology. Till now, we have identified key pathways that are activated and showed that most of these pathway activations are due to downregulation of regulating miRNAs. The downregulation of these miRNAs are mediated by EBV infection or MYC activation. We have also identified certain markers that are universally over-expressed in NKTL and may serve as novel therapeutic targets.