
Yilong ZHOU
RNA is continuously transcribed, processed, exported, translated, and degraded within cells, a dynamic process shaped by various RNA-binding proteins, membrane organelles, and biomolecular condensates—like a river shaped by rocks, banks, and lakes. Our lab investigates the dynamics of RNA flux, focusing on understanding RNA-Protein interactions and phase separation, RNA modifications and splicing changes under stress and disease conditions. We aim to uncover RNA-related disease mechanisms and identify therapeutic RNA targets. Our approach spans molecular to animal models, integrating advanced technologies such as deep sequencing, mass spectrometry, phase separation assays, iCLIP, and our self-developed "FANCI" method.
zhou@nus.edu.sg
Principal Investigator, Cancer Science Institute of Singapore
Assistant Professor, Department of Biochemistry, Yong Loo Lin School of Medicine, NUS
| 2025 | Presidential Young Professorship, National University of Singapore |
| 2018 | EMBO Long-Term Fellowship |
Our research focuses on understanding how cells respond to dangerous RNA species, including endogenous damaged RNA and exogenous virus RNA, under different stress and pathological conditions.
We developed a new method called “FANCI” and identified a unique subtype of RNA condensates known as DHX9 stress granules (SGs) in the cells. These DHX9 SGs sequester damaged intron RNA in the cytosol and activate a series of protective stress responses such as inflammation and P62 autophagy as well as protein translation shutdown. Their assembly represents a novel cytoprotective mechanism in cells, safeguarding them from RNA damage (Figure 1, left panel).
Moreover, we identified two critical but distinct regulators—CSNK1E (a kinase) and PLCB2 (a phospholipase)—that control immune responses to virus dsRNA. Using cell and animal models, along with patient-derived samples, we revealed their physiological roles and underlying molecular mechanisms (Figure 1, right panel).
In the future, we aim to further investigate the principles and molecular mechanisms underlying “toxic” RNA response, with the goal of using this knowledge to gain insights into RNA-related disease mechanisms and identify potential therapeutic targets.
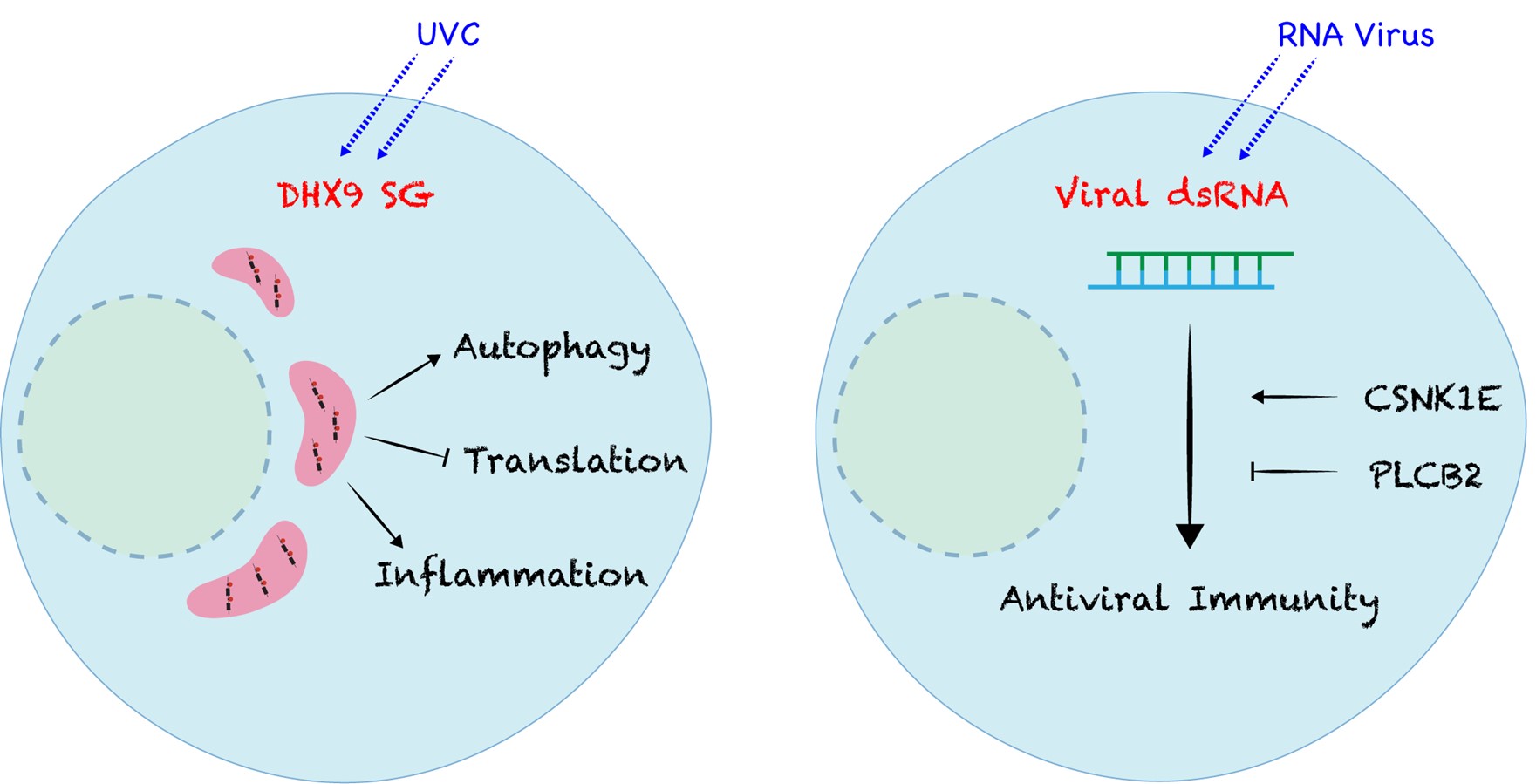
Figure 1: Cellular responses to “toxic” RNA species
1. Zhou, Y., Panhale, A., Shvedunova, M., Balan, M., Gomez-Auli, A., Holz, H., Seyfferth, J., Helmstädter, M., Kayser, S., Zhao, Y., Erdogdu, N. U., Grzadzielewska, I., Mittler, G., Manke, T., & Akhtar, A. (2024). RNA damage compartmentalization by DHX9 stress granules. Cell. 187(7):1701-1718.e28. https://pubmed.ncbi.nlm.nih.gov/38503283/
2. Wang, F., Zhao, M., Chang, B., Zhou, Y., Wu, X., Ma, M., Liu, S., Cao, Y., Zheng, M., Dang, Y., Xu, J., Chen, L., Liu, T., Tang, F., Ren, Y., Xu, Z., Mao, Z., Huang, K., Luo, M., … Ge, B. (2022). Cytoplasmic PARP1 links the genome instability to the inhibition of antiviral immunity through PARylating cGAS. Molecular Cell, 82(11), 2032–2049.e7. https://pubmed.ncbi.nlm.nih.gov/35460603/
3. Wang, L., Liu, Z., Wang, J., Liu, H., Wu, J., Tang, T., Li, H., Yang, H., Qin, L., Ma, D., Chen, J., Liu, F., Wang, P., Zheng, R., Song, P., Zhou, Y., Cui, Z., Wu, X., Huang, X., … Ge, B. (2019). Oxidization of TGFβ-activated kinase by MPT53 is required for immunity to Mycobacterium tuberculosis. Nature Microbiology, 4(8), 1378–1388. https://pubmed.ncbi.nlm.nih.gov/31110366/
4. Wang, L.*, Zhou, Y.*, Chen, Z.*, Sun, L., Wu, J., Li, H., Liu, F., Wang, F., Yang, C., Yang, J., Leng, Q., Zhang, Q., Xu, A., Shen, L., Sun, J., Wu, D., Fang, C., Lu, H., Yan, D., & Ge, B. (2019). PLCβ2 negatively regulates the inflammatory response to virus infection by inhibiting phosphoinositide-mediated activation of TAK1. Nature Communications, 10(1), 746. (co-first author) https://pubmed.ncbi.nlm.nih.gov/30765691/
5. Liu, F., Chen, J., Wang, P., Li, H., Zhou, Y., Liu, H., Liu, Z., Zheng, R., Wang, L., Yang, H., Cui, Z., Wang, F., Huang, X., Wang, J., Sha, W., Xiao, H., & Ge, B. (2018). MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy. Nature Communications, 9(1), 4295. https://pubmed.ncbi.nlm.nih.gov/30327467/
6. Zheng, R., Liu, H., Zhou, Y., Yan, D., Chen, J., Ma, D., Feng, Y., Qin, L., Liu, F., Huang, X., Wang, J., & Ge, B. (2018). Notch4 Negatively Regulates the Inflammatory Response to Mycobacterium tuberculosis Infection by Inhibiting TAK1 Activation. The Journal of Infectious Diseases, 218(2), 312–323. https://pubmed.ncbi.nlm.nih.gov/29228365/
7. He, C., Zhou, Y., Liu, F., Liu, H., Tan, H., Jin, S., Wu, W., & Ge, B. (2017). Bacterial Nucleotidyl Cyclase Inhibits the Host Innate Immune Response by Suppressing TAK1 Activation. Infection and Immunity, 85(9). https://pubmed.ncbi.nlm.nih.gov/28652310/
8. Zhou, Y., He, C., Wang, L., & Ge, B. (2017). Post-translational regulation of antiviral innate signaling. European Journal of Immunology, 47(9), 1414–1426. https://pubmed.ncbi.nlm.nih.gov/28744851/
9. Zhou, Y., He, C., Yan, D., Liu, F., Liu, H., Chen, J., Cao, T., Zuo, M., Wang, P., Ge, Y., Lu, H., Tong, Q., Qin, C., Deng, Y., & Ge, B. (2016). The kinase CK1ɛ controls the antiviral immune response by phosphorylating the signaling adaptor TRAF3. Nature Immunology, 17(4), 397–405. https://pubmed.ncbi.nlm.nih.gov/26928339/
10. Bai, W., Liu, H., Ji, Q., Zhou, Y., Liang, L., Zheng, R., Chen, J., Liu, Z., Yang, H., Zhang, P., Kaufmann, S. H. E., & Ge, B. (2014). TLR3 regulates mycobacterial RNA-induced IL-10 production through the PI3K/AKT signaling pathway. Cellular Signalling, 26(5), 942–950. https://pubmed.ncbi.nlm.nih.gov/24462705/





