
Polly Leilei CHEN
The human genome encodes RNA molecules that instruct the behaviour of cancer cells. Leveraging on the next-generation sequencing technologies, well-established cancer cell lines and animal models, A/Prof. Polly Leilei Chen’s group aims to identify the harmful changes that will occur in RNA molecules after it is made from DNA. It is critical to uncover how these changes trigger a cell to develop into cancer which can eventually lead to the unsatisfactory treatment outcomes for patients with cancers (particularly esophageal, stomach, colon and liver cancer). The long-term goal of A/Prof. Chen’s team is to develop novel cancer therapies targeting these cancer-associated RNA changes.
polly_chen[at]nus.edu.sg
Principal Investigator, Cancer Science Institute of Singapore
Associate Professor, Department of Anatomy, NUS
| 2023 | Yong Loo Lin School of Medicine Research of The Year Award 2023 |
| 2021 | Yong Loo Lin School of Medicine Research Excellence Award |
| 2019 | EMBO Young Investigator Program (YIP) |
| 2018 | NUHS-Mochtar Riady Pinnacle Awards-High Achiever Award 2018 |
| 2017 | Yong Loo Lin School of Medicine Young Researcher Of the Year Award |
| 2015 | President Assistant Professorship, National University of Singapore |
| 2014 | Science and Technology Award of Higher Education of China (Second Class), Ministry of Education, China |
| 2014 | NUS Young Scientist Award, National University of Singapore |
| 2010 | Li Ka Shing Prize 2009/2010, Hong Kong |
My research interest has been centered on understanding the changes in RNA (particularly adenosine-to-inosine RNA editing and splicing) in human cancers and translating this new knowledge to aid early detection and intervention as well as treatment.
RNA editing and splicing share the largest overlap in time and space during processing of pre-mRNA and contribute to transcriptome diversity. RNA editing introduces changes in RNA sequences encoded by the genome. In humans, the most common type of editing is the conversion of adenosine to inosine (A-to-I), which is catalysed by the double-stranded RNA-specific adenosine deaminase that acts on RNA (ADAR) family of proteins. This process of recoding of translated exons, widespread editing of repetitive sequence elements and sequence modification of microRNA (miRNA) can lead to specific amino acid substitutions, alternative splicing, and changes in gene expression levels. With the advent of high-throughput transcriptome sequencing, many editing sites potentially associated with cancer development have been identified.
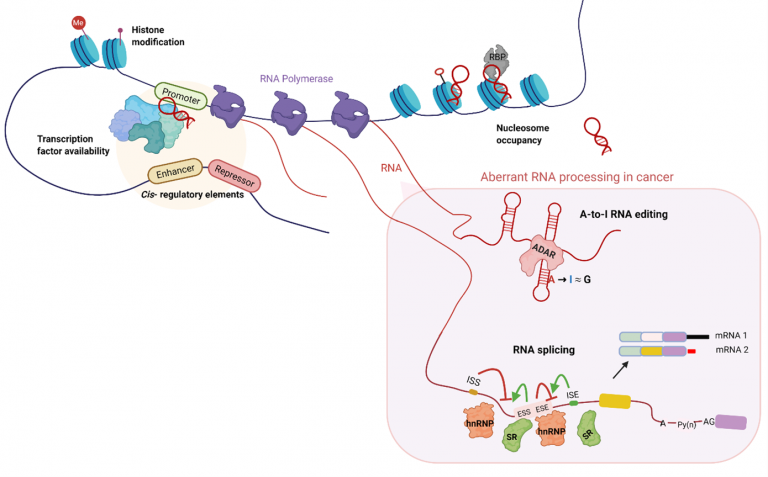
| Alternative splicing and RNA editing are the two major pre-messenger RNA processing steps diversifying transcriptome in eukaryotes. ADAR1 and/or ADAR2 proteins bind to double-stranded RNAs (dsRNAs) in the nucleus and convert adenosine (A) to inosine (I). Inosine is recognized as guanosine (G) by most cellular machineries, such as splicing and translation machineries. Regulation of alternative splicing involves cis-acting elements, such as exonic/intronic splicing enhancers/silencers (ESE/S, ISE/S), and transacting factors, such as serine arginine-rich (SR) proteins and heterogeneous nuclear ribonucleoproteins (hnRNPs), transcription, chromatin modification, and RNA secondary structure. |
We were amongst the first to report that the dysregulated adenosine-to-inosine (A-to-I) RNA editing contributes to human liver, stomach, and oesophageal cancers. We deciphered how RNA editing enzyme ADARs regulate gene functions and other RNA processing steps (e.g., canonical linear mRNA splicing and non-canonical backsplicing) via their editing-dependent and/or independent functions in the context of cancer and identified hundreds of non-ADAR editing regulators and decipher their role in shaping the cancer editome. To translate our research into clinically applied settings, we developed antisense oligonucleotide (ASO)-based RNA editing inhibitors for cancer treatment (International Patent Application No. PCT/SG2020/050380) and identified a 29-gene RNA editing signature for guiding gastric cancer chemotherapy (International Patent Application No. PCT/SG2021/050203).
My recent research focuses have been placed on 1) identifying changes in RNAs leading to cancer initiation, relapse post treatment, and drug resistance using advanced multi-omics approaches; and 2) understanding how pre-malignant and tumour cells develop immune evasion mechanisms through impairing RNA sensing and immune activation.
- Han J, Song Y, Xie J, Tano V, Shen H, Gan WL, Ng L, Ng BYL, Ng VHE, Sui X, Tang SJ and Chen L. Modulation of N6-methyladenosine (m6A) RNA Modification by Death Associated Protein 3 (DAP3) in Cancer Cells. Proc Natl Acad Sci U S A, 2024, 121 (40), e2404509121.
- Gan WL, Ren X, Ng VHE, Ng L, Song Y, Tano V, Han J, An O, Xie J, Ng BYL, Tay DJT, Tang SJ, Shen H, Khare S, Chong KHC, Young DY, Wu B, DasGupta R, and Chen L. Hepatocyte-macrophage crosstalk via the PGRN-EGFR axis modulates ADAR1-mediated immunity in the liver. Cell Reports, 2024, 43, 114400.
- Guo M, Chan TH, Zhou Q, An Ő, Li Y, Song Y, Tan HZ, Ng VHE, Peramangalam PS, Tan ZQ, Cao X, Iwanaga E, Matsuoka M, Ooi M, Jen WY, Koh LP, Chan E, Tan LK, Goh Y, Wang W, Koh BTH, Chan MC, Fullwood MJ, Chng WJ, Osato M, Pulikkan JA, Yang H, Chen L*, Tenen DG*. Core binding factor fusion downregulation of ADAR2 RNA editing contributes to AML leukemogenesis. Blood, 2023 Feb 16; blood.2022015830. (*, co-last)
- Chen L. A-to-I editing prevents self-RNA sensing. Nature Reviews Molecular Cell Biology (2022). https://doi.org/10.1038/s41580-022-00540-4
- Shen H, An Ö, Ren X, Song Y, Tang SJ, Ke X, Han J, Tay DJ, Ng VHE, Bellido Molias F, Pitcheshwar P, Leong KW, Tan KK , Yang H, Chen L. ADARs act as potent regulators of circular transcriptome in cancer. Nature Communications, 13(1):16 pages Number ARTN 1508 21 Mar 2022.
- Tay DJ, Song Y, Peng B, Toh TB, Hooi L, Toh DFK, Hong H, Tang SJ, Han J, Gan WL, Chan THM, Krishna MS, Patil KM, Maraswami M, Loh TP, Dan YY, Zhou L, Bonney GK, Chow PKH, Chen G, Chow EKH, Le MT, Chen L. Targeting RNA Editing of Antizyme Inhibitor 1: a Potential Oligonucleotide-Based Antisense Therapy for Cancer. Molecular Therapy, 2021, 29(11):3258-3273.
- Song Y, An O, Ren X, Chan TH, Tay DJ, Tang SJ, Han J, Hong HQ, Ng VHE, Ke X, Shen H, Pitcheshwar P, Lin SJ, Leong KW, Molias FB, Yang H, Kappei D, Chen L. RNA editing mediates the functional switch of COPA in a novel mechanism of hepatocarcinogenesis. Journal of Hepatology. Published: July 18, 2020. DOI:https://doi.org/10.1016/j.jhep.2020.07.021
- Han J, An O, Hong H, Chan TH, Song Y, Shen H, Tang SJ, Lin JS, Ng VHE, Tay DJ, Molias FB, Pitcheshwar P, Yang H, Chen L. Suppression of Adenosine-to-Inosine (A-to-I) RNA Editome by Death Associated Protein 3 (DAP3) Promotes Cancer Progression. Science Advances,2020; 6, eaba5136.
- Tang SJ, Shen H, An O, Hong HQ, Li J, Song Y, Han J, Tay DJ, Ng VHE, Molias FB, Leong KW, Pitcheshwar P, Yang H, Chen L. Cis- and trans-regulation of pre-mRNA splicing by RNA editing enzymes: influencing cancer development. Nature Communications. 2020;11(1):799.
- Hong H, An O, Chan TH, Ng VHE, Kwok HS, Lin JS, Qi L, Han J, Tay DJ, Tang SJ, Yang H, Song YY, Molias FB, Tenen DG, Chen L. Bidirectional regulation of adenosine-to-inosine (A-to-I) RNA editing by DEAH box helicase 9 (DHX9) in cancer. Nucleic Acids Res, 2018 Sep 6;46(15):7953-7969.















