Quantitative Proteomics Core
The Quantitative Proteomics Core at the Cancer Science Institute of Singapore offers a complete package for proteomics applications all the way from help with experimental design over sample preparation and mass spectrometry measurement to data analysis, visualization and storage. We cover a broad range of proteomics applications including clinical samples.
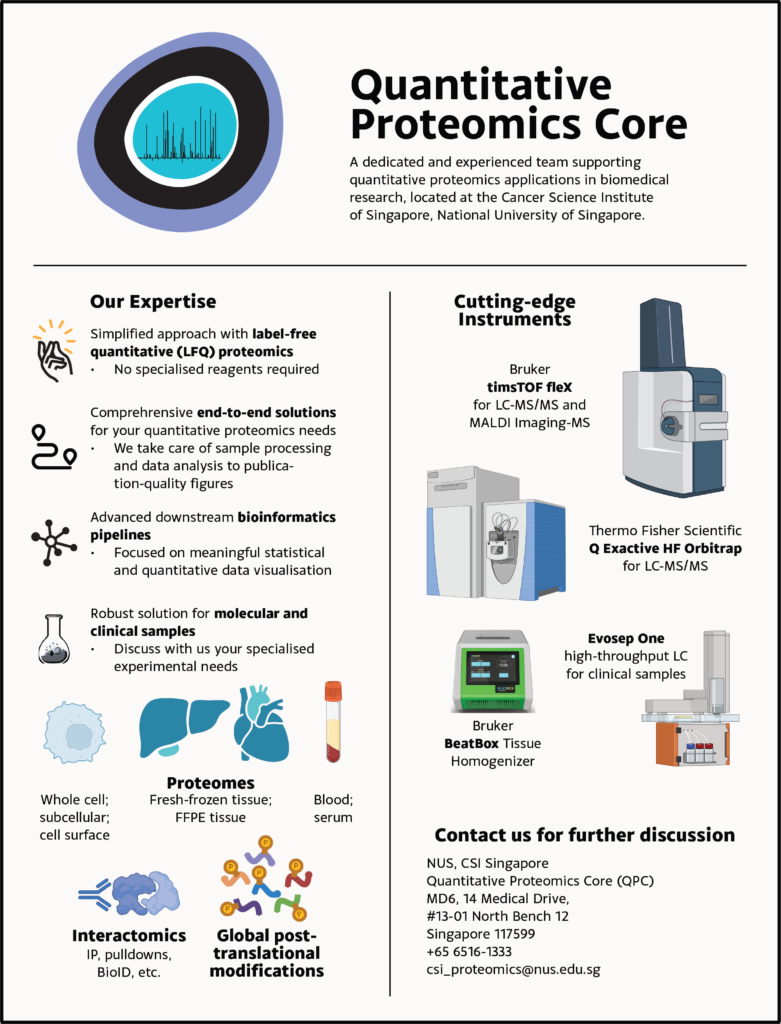
We currently operate a Q Exactive HF hybrid quadrupole-Orbitrap mass spectrometer as well as a timsTOF flex in combination with an EASY-nLC 1200 uHPLC system or an Evosep ONE. The data analysis is performed on in-house servers using the freely available quantitative proteomics software package MaxQuant. We are committed to providing quantitative mass spectrometry solutions and to this end, support researchers in using SILAC (Stable Isotope Labeling by Amino Acids in Cell Culture) or label-free quantitation (LFQ).
For further information and to discuss how we may help you with your specific experimental questions, please contact the Quantitative Proteomics Core via mail (csi_proteomics[at]nus.edu.sg).
Facility Head

Dr. Dennis KAPPEI
Dennis’ research group uses their expertise in quantitative proteomics to study chromatin changes in the context of telomere-driven genome instability. Through the Quantitative Proteomics Core, this know-how in mass spectrometry applications is shared with CSI and the wider Singapore research community.
Team





Dr Wai Khang YONG
Core Facility Manager
Wai Khang earned a PhD from NUS Graduate School's Integrative Sciences and Engineering Programme (ISEP). His research centers on investigating chromatin protein-DNA interactions through ChIP-MS. With meticulous attention to technical nuances, he has implemented in-house experimental pipelines for ChIP-MS, surface membrane proteomics, and PTM enrichment.
Dr Sakshi RAJORIA
Research Fellow
After graduate training at the Defence Research and Development Establishment in Madhya Pradesh, Sakshi spent initial postdoctoral stints at IIT Bombay and the University of Massachusetts. In our team she is developing spatial proteomics workflows to analyse tissue samples by imaging mass spectrometry.
Vartika KHANCHANDANI
Research Assistant
Vartika earned a Bachelor of Engineering from the R.V. College of Engineering in Bangalore. In our group, she is applying her bioinformatics skills to the development of streamlined analysis pipelines.
Jie Min LEE
Research Assistant
Jie Min earned her Bachelor of Science in Microbiology from the University of Malaya. As part of the QPC, she studies protein expression and interactions in various biological contexts. Her role encompasses different aspects of mass spectrometry, including sample preparation, instrument operation, maintenance and data analysis.
Sumedha PUNDRIK
Research Assistant
Sumedha obtained her MSc in Precision Medicine from the University of Manchester. Within the team, she manages tasks related to sample preparation and data analysis, focusing on investigating protein expression and interactions across diverse biological contexts.

